Genomic diversity of SNPs across Anolis carolinensis lizard populations
/A fun paper, lead by Yann Bourgeois, was just published in Genome Biology and Evolution. It describes the variation in nucleotide polymorphisms across five major populations of green anoles in the southern US, their likely demographic changes across time, and evolutionary dynamics that have likely shaped the mutation composition of their chromosomes. Congratulations, Yann!
Recent secondary contacts, linked selection and variable recombination rates shape genomic diversity in the model species Anolis carolinensis.
Y Bourgeois, RP Ruggiero, JD Manthey, & S Boissinot (2019) Genome Biology and Evolution
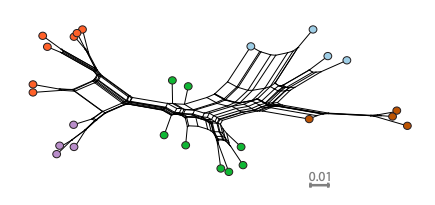
Gaining a better understanding on how selection and neutral processes affect genomic diversity is essential to gain better insights into the mechanisms driving adaptation and speciation. However, the evolutionary processes affecting variation at a genomic scale have not been investigated in most vertebrate lineages. Here we present the first population genomics survey using whole genome re-sequencing in the green anole (Anolis carolinensis). Anoles have been intensively studied to understand mechanisms underlying adaptation and speciation. The green anole in particular is an important model to study genome evolution. We quantified how demography, recombination and selection have led to the current genetic diversity of the green anole by using whole-genome resequencing of five genetic clusters covering the entire species range. The differentiation of green anole’s populations is consistent with a northward expansion from South Florida followed by genetic isolation and subsequent gene flow among adjacent genetic clusters. Dispersal out-of-Florida was accompanied by a drastic population bottleneck followed by a rapid population expansion. This event was accompanied by male-biased dispersal and/or selective sweeps on the X chromosome. We show that the interaction between linked selection and recombination is the main contributor to the genomic landscape of differentiation in the anole genome.